Overview
What is Intake-ESM?
How can we use it to read in data?
How do we work with dictionaries of datasets?
How do I write my analysis to work with Intake-ESM?
Setup
Imports
Here, we will import intake, and dask.distributed (distributed)
Remember, Intake-ESM is a plugin within the Intake project, so we do not need to explicitly call
import intake_esm
import intake
from distributed import Client, LocalCluster
import xarray as xr
import matplotlib.pyplot as plt
import fsspec
Spin up a Dask Cluster
We will go ahead and spin up a Dask Cluster
cluster = LocalCluster()
client = Client(cluster)
client
/Users/mgrover/miniforge3/envs/mscar-python-tutorial-dev/lib/python3.10/site-packages/distributed/node.py:183: UserWarning: Port 8787 is already in use.
Perhaps you already have a cluster running?
Hosting the HTTP server on port 57407 instead
warnings.warn(
Client
Client-c8ff070e-420f-11ed-a074-520a01803a93
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:57407/status |
Cluster Info
LocalCluster
7c72d1a8
| Dashboard: http://127.0.0.1:57407/status | Workers: 5 |
| Total threads: 10 | Total memory: 32.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-9e871823-4c3b-4f19-9efa-61d916417a76
| Comm: tcp://127.0.0.1:57408 | Workers: 5 |
| Dashboard: http://127.0.0.1:57407/status | Total threads: 10 |
| Started: Just now | Total memory: 32.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:57432 | Total threads: 2 |
| Dashboard: http://127.0.0.1:57433/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:57413 | |
| Local directory: /var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/dask-worker-space/worker-c789voz8 | |
Worker: 1
| Comm: tcp://127.0.0.1:57427 | Total threads: 2 |
| Dashboard: http://127.0.0.1:57429/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:57412 | |
| Local directory: /var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/dask-worker-space/worker-yiv0bk22 | |
Worker: 2
| Comm: tcp://127.0.0.1:57435 | Total threads: 2 |
| Dashboard: http://127.0.0.1:57437/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:57415 | |
| Local directory: /var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/dask-worker-space/worker-hetk3utz | |
Worker: 3
| Comm: tcp://127.0.0.1:57426 | Total threads: 2 |
| Dashboard: http://127.0.0.1:57428/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:57411 | |
| Local directory: /var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/dask-worker-space/worker-ef2rd9n4 | |
Worker: 4
| Comm: tcp://127.0.0.1:57436 | Total threads: 2 |
| Dashboard: http://127.0.0.1:57439/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:57414 | |
| Local directory: /var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/dask-worker-space/worker-ivhltpnd | |
How can I use Intake-ESM to read in my data?
“Traditional” Workflow
Say, for example, wanted to take a look at data from the AWS hosted Community Earth System Model Large Ensemble (CESM-LENS)…
We want to take a look at:
Atmospheric temperature (
T)Atmospheric moisture (
Q)
As well as:
Ocean temperature (
TEMP)Ocean salinity (
SALT)
Investigate the files on AWS
We use fsspec here to take a look at what files are available; this would be similar to listing files on your local system (ex. glob)
fs = fsspec.filesystem('s3', anon=True)
bucket = 'ncar-cesm-lens'
fs.ls(bucket)
['ncar-cesm-lens/atm',
'ncar-cesm-lens/catalogs',
'ncar-cesm-lens/ice_nh',
'ncar-cesm-lens/ice_sh',
'ncar-cesm-lens/lnd',
'ncar-cesm-lens/ocn']
You’ll notice we have a few directories in this bucket, corresponding to each component:
Atmosphere (
atm)Ice Northern Hemisphere (
ice_nh)Ice Southern Hemisphere (
ice_sh)Land (
lnd)Ocean (
ocn)
If we go into the atm directory, we see there are various frequencies which each component as well
bucket = 'ncar-cesm-lens/atm'
fs.ls(bucket)
['ncar-cesm-lens/atm/',
'ncar-cesm-lens/atm/daily',
'ncar-cesm-lens/atm/hourly6-1990-2005',
'ncar-cesm-lens/atm/hourly6-2026-2035',
'ncar-cesm-lens/atm/hourly6-2071-2080',
'ncar-cesm-lens/atm/monthly',
'ncar-cesm-lens/atm/static']
If we go one level further, we see the actual data, separated by cesmLE-{experiment}-{variable}.zarr
bucket = 'ncar-cesm-lens/atm/monthly'
fs.ls(bucket)
['ncar-cesm-lens/atm/monthly/',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-FLNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-FLNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-FLUT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-FSNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-FSNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-FSNTOA.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-ICEFRAC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-LHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-PRECC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-PRECL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-PRECSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-PRECSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-PSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-Q.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-SHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-T.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-TMQ.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-TREFHT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-TREFHTMN.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-TREFHTMX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-TS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-U.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-V.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-20C-Z3.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-FLNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-FLNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-FLUT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-FSNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-FSNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-FSNTOA.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-ICEFRAC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-LHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-PRECC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-PRECL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-PRECSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-PRECSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-PS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-PSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-Q.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-SHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-T.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-TMQ.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-TREFHT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-TREFHTMN.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-TREFHTMX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-TS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-U.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-V.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-CTRL-Z3.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-FLNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-FLNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-FLUT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-FSNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-FSNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-FSNTOA.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-ICEFRAC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-LHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-PRECC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-PRECL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-PRECSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-PRECSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-PSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-Q.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-SHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-T.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-TMQ.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-TREFHT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-TREFHTMN.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-TREFHTMX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-TS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-U.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-V.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-HIST-Z3.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-FLNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-FLNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-FLUT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-FSNS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-FSNSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-FSNTOA.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-ICEFRAC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-LHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-PRECC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-PRECL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-PRECSC.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-PRECSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-PS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-PSL.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-Q.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-SHFLX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-T.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-TMQ.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-TREFHT.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-TREFHTMN.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-TREFHTMX.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-TS.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-U.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-V.zarr',
'ncar-cesm-lens/atm/monthly/cesmLE-RCP85-Z3.zarr']
Loading in Data Using Xarray
Let’s say we wanted to look at data from the historical scenario (HIST)… We could load the data using the following syntax!
var = 'T'
atmosphere_ds = xr.open_zarr(f's3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-{var}.zarr', storage_options={'anon':True})
atmosphere_ds
<xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes:
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
host: tcs-f02n07
important_note: This data is part of the project 'Blind Evalua...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.105.cam.i.2006-01-...
logname: mudryk
nco_openmp_thread_number: 1
revision_Id: $Id$
source: CAM
title: UNSET
topography_file: /scratch/p/pjk/mudryk/cesm1_1_2_LENS/inputdata...If we wanted all of these in the same dataset, we would need to write the merging ourselves
variables = ['T', 'Q']
ds_list = []
# Loop through the different files and add them to the list of datasets
for var in variables:
ds_list.append(xr.open_zarr(f's3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-{var}.zarr', storage_options={'anon':True}))
atmosphere_merged = xr.merge(ds_list)
atmosphere_merged
<xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes:
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
host: tcs-f02n07
important_note: This data is part of the project 'Blind Evalua...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.105.cam.i.2006-01-...
logname: mudryk
nco_openmp_thread_number: 1
revision_Id: $Id$
source: CAM
title: UNSET
topography_file: /scratch/p/pjk/mudryk/cesm1_1_2_LENS/inputdata...That wasn’t too bad, what if we wanted to look look at other experiments? Or other components, such as the ocean? It gets tricky…
Experiments have different time ranges
Components have different grids
Let’s load in an ocean temperature dataset…
ocean_ds = xr.open_zarr(f's3://ncar-cesm-lens/ocn/monthly/cesmLE-RCP85-TEMP.zarr', storage_options={'anon':True})
ocean_ds
<xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, z_t: 60, nlat: 384, nlon: 320, d2: 2)
Coordinates:
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bound (time, d2) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
* z_t (z_t) float32 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
Dimensions without coordinates: nlat, nlon, d2
Data variables:
TEMP (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
Attributes:
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/eaton/netc...
NCO: 4.3.4
calendar: All years have exactly 365 days.
cell_methods: cell_methods = time: mean ==> the variable val...
contents: Diagnostic and Prognostic Variables
nco_openmp_thread_number: 1
nsteps_total: 750
revision: $Id: tavg.F90 41939 2012-11-14 16:37:23Z mlevy...
source: CCSM POP2, the CCSM Ocean Component
start_time: This dataset was created on 2014-12-26 at 15:5...
tavg_sum: 2592000.0
tavg_sum_qflux: 2592000.0Notice how we now have dimensions (time, z_t, nlat, nlon), which differs from the atmosphere dataset (time, lev, lat, lon)
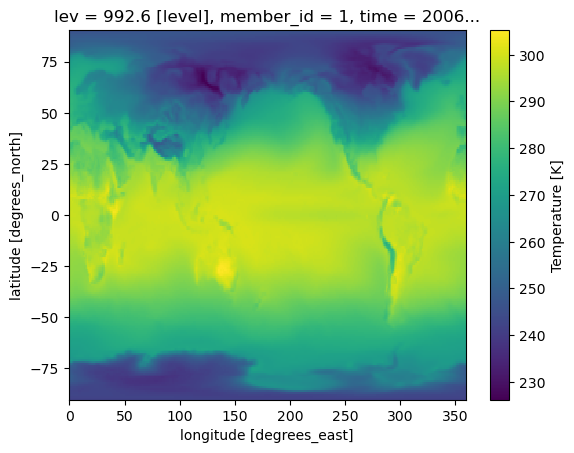
atmosphere_ds.isel(member_id=0, time=0, lev=-1).T.plot();
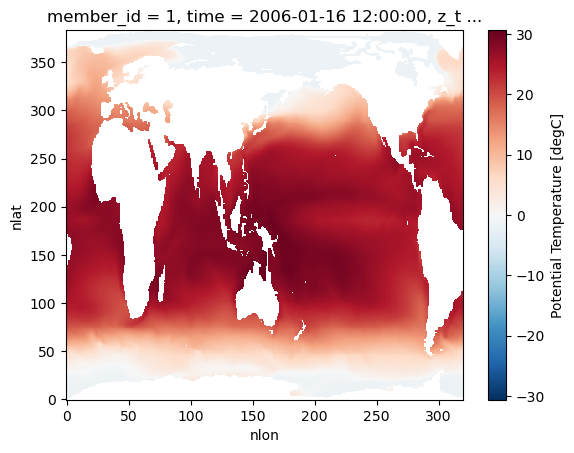
ocean_ds.isel(member_id=0, time=0, z_t=0).TEMP.plot();
What if there were an easier way of searching for available data, as well as loading the data into your analysis to make it easier to generalize? This is where Intake-ESM comes in!
Intake-ESM Method
We can use the Intake-ESM catalog from the CESM-LENS dataset (https://raw.githubusercontent.com/NCAR/cesm-lens-aws/master/intake-catalogs/aws-cesm1-le.json) to work with these data!
data_catalog = intake.open_esm_datastore('https://raw.githubusercontent.com/NCAR/cesm-lens-aws/master/intake-catalogs/aws-cesm1-le.json')
data_catalog
aws-cesm1-le catalog with 56 dataset(s) from 442 asset(s):
| unique | |
|---|---|
| variable | 78 |
| long_name | 75 |
| component | 5 |
| experiment | 4 |
| frequency | 6 |
| vertical_levels | 3 |
| spatial_domain | 5 |
| units | 25 |
| start_time | 12 |
| end_time | 13 |
| path | 427 |
| derived_variable | 0 |
You’ll notice that this catalog has 5 components, and six frequencies as we mentioned earlier. If we call .df on this catalog, we can see the table of metadata!
data_catalog.df
| variable | long_name | component | experiment | frequency | vertical_levels | spatial_domain | units | start_time | end_time | path | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | FLNS | net longwave flux at surface | atm | 20C | daily | 1.0 | global | W/m2 | 1920-01-01 12:00:00 | 2005-12-31 12:00:00 | s3://ncar-cesm-lens/atm/daily/cesmLE-20C-FLNS.... |
| 1 | FLNSC | clearsky net longwave flux at surface | atm | 20C | daily | 1.0 | global | W/m2 | 1920-01-01 12:00:00 | 2005-12-31 12:00:00 | s3://ncar-cesm-lens/atm/daily/cesmLE-20C-FLNSC... |
| 2 | FLUT | upwelling longwave flux at top of model | atm | 20C | daily | 1.0 | global | W/m2 | 1920-01-01 12:00:00 | 2005-12-31 12:00:00 | s3://ncar-cesm-lens/atm/daily/cesmLE-20C-FLUT.... |
| 3 | FSNS | net solar flux at surface | atm | 20C | daily | 1.0 | global | W/m2 | 1920-01-01 12:00:00 | 2005-12-31 12:00:00 | s3://ncar-cesm-lens/atm/daily/cesmLE-20C-FSNS.... |
| 4 | FSNSC | clearsky net solar flux at surface | atm | 20C | daily | 1.0 | global | W/m2 | 1920-01-01 12:00:00 | 2005-12-31 12:00:00 | s3://ncar-cesm-lens/atm/daily/cesmLE-20C-FSNSC... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 437 | WVEL | vertical velocity | ocn | RCP85 | monthly | 60.0 | global_ocean | centimeter/s | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-RCP85-W... |
| 438 | NaN | NaN | ocn | CTRL | static | NaN | global_ocean | NaN | NaN | NaN | s3://ncar-cesm-lens/ocn/static/grid.zarr |
| 439 | NaN | NaN | ocn | HIST | static | NaN | global_ocean | NaN | NaN | NaN | s3://ncar-cesm-lens/ocn/static/grid.zarr |
| 440 | NaN | NaN | ocn | RCP85 | static | NaN | global_ocean | NaN | NaN | NaN | s3://ncar-cesm-lens/ocn/static/grid.zarr |
| 441 | NaN | NaN | ocn | 20C | static | NaN | global_ocean | NaN | NaN | NaN | s3://ncar-cesm-lens/ocn/static/grid.zarr |
442 rows × 11 columns
Searching for your Data
We can use the .search API to search for the data we are interested in. In this case, as mentioned before, we are interested in future data (RCP85) from the atmosphere and ocean
data_catalog_subset = data_catalog.search(variable=['T', 'Q', 'TEMP', 'SALT'],
frequency='monthly',)
data_catalog_subset.df
| variable | long_name | component | experiment | frequency | vertical_levels | spatial_domain | units | start_time | end_time | path | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Q | specific humidity | atm | 20C | monthly | 30.0 | global | kg/kg | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-20C-Q.zarr |
| 1 | T | temperature | atm | 20C | monthly | 30.0 | global | K | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-20C-T.zarr |
| 2 | Q | specific humidity | atm | CTRL | monthly | 30.0 | global | kg/kg | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-CTRL-Q.... |
| 3 | T | temperature | atm | CTRL | monthly | 30.0 | global | K | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-CTRL-T.... |
| 4 | Q | specific humidity | atm | HIST | monthly | 30.0 | global | kg/kg | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-HIST-Q.... |
| 5 | T | temperature | atm | HIST | monthly | 30.0 | global | K | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-HIST-T.... |
| 6 | Q | specific humidity | atm | RCP85 | monthly | 30.0 | global | kg/kg | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-Q... |
| 7 | T | temperature | atm | RCP85 | monthly | 30.0 | global | K | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-T... |
| 8 | SALT | salinity | ocn | 20C | monthly | 60.0 | global_ocean | gram/kilogram | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-20C-SAL... |
| 9 | TEMP | potential temperature | ocn | 20C | monthly | 60.0 | global_ocean | degC | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-20C-TEM... |
| 10 | SALT | salinity | ocn | CTRL | monthly | 60.0 | global_ocean | gram/kilogram | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-CTRL-SA... |
| 11 | TEMP | potential temperature | ocn | CTRL | monthly | 60.0 | global_ocean | degC | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-CTRL-TE... |
| 12 | SALT | salinity | ocn | HIST | monthly | 60.0 | global_ocean | gram/kilogram | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-HIST-SA... |
| 13 | TEMP | potential temperature | ocn | HIST | monthly | 60.0 | global_ocean | degC | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-HIST-TE... |
| 14 | SALT | salinity | ocn | RCP85 | monthly | 60.0 | global_ocean | gram/kilogram | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-RCP85-S... |
| 15 | TEMP | potential temperature | ocn | RCP85 | monthly | 60.0 | global_ocean | degC | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-RCP85-T... |
We can take a look at our catalog dataframe again, to verify we have the datasets we are looking for!
data_catalog_subset.df
| variable | long_name | component | experiment | frequency | vertical_levels | spatial_domain | units | start_time | end_time | path | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Q | specific humidity | atm | 20C | monthly | 30.0 | global | kg/kg | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-20C-Q.zarr |
| 1 | T | temperature | atm | 20C | monthly | 30.0 | global | K | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-20C-T.zarr |
| 2 | Q | specific humidity | atm | CTRL | monthly | 30.0 | global | kg/kg | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-CTRL-Q.... |
| 3 | T | temperature | atm | CTRL | monthly | 30.0 | global | K | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-CTRL-T.... |
| 4 | Q | specific humidity | atm | HIST | monthly | 30.0 | global | kg/kg | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-HIST-Q.... |
| 5 | T | temperature | atm | HIST | monthly | 30.0 | global | K | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-HIST-T.... |
| 6 | Q | specific humidity | atm | RCP85 | monthly | 30.0 | global | kg/kg | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-Q... |
| 7 | T | temperature | atm | RCP85 | monthly | 30.0 | global | K | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-T... |
| 8 | SALT | salinity | ocn | 20C | monthly | 60.0 | global_ocean | gram/kilogram | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-20C-SAL... |
| 9 | TEMP | potential temperature | ocn | 20C | monthly | 60.0 | global_ocean | degC | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-20C-TEM... |
| 10 | SALT | salinity | ocn | CTRL | monthly | 60.0 | global_ocean | gram/kilogram | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-CTRL-SA... |
| 11 | TEMP | potential temperature | ocn | CTRL | monthly | 60.0 | global_ocean | degC | 0400-01-16 12:00:00 | 2200-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-CTRL-TE... |
| 12 | SALT | salinity | ocn | HIST | monthly | 60.0 | global_ocean | gram/kilogram | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-HIST-SA... |
| 13 | TEMP | potential temperature | ocn | HIST | monthly | 60.0 | global_ocean | degC | 1850-01-16 12:00:00 | 1919-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-HIST-TE... |
| 14 | SALT | salinity | ocn | RCP85 | monthly | 60.0 | global_ocean | gram/kilogram | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-RCP85-S... |
| 15 | TEMP | potential temperature | ocn | RCP85 | monthly | 60.0 | global_ocean | degC | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/ocn/monthly/cesmLE-RCP85-T... |
Load in the Data
Now that we have subset our catalog, we can load the data into our notebook using .to_dataset_dict() which is short for “to dataset dictionary”
Intake-ESM aggregates these datasets by component, experiment, and frequency, and provides a nice progress bar to check in on the data access process!
dsets = data_catalog_subset.to_dataset_dict(storage_options={'anon':True})
dsets
--> The keys in the returned dictionary of datasets are constructed as follows:
'component.experiment.frequency'
{'ocn.20C.monthly': <xarray.Dataset>
Dimensions: (member_id: 40, time: 1032, z_t: 60, nlat: 384, nlon: 320, d2: 2)
Coordinates:
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 1920-01-16 12:00:00 ... 2005-12-16 12:00:00
time_bound (time, d2) object dask.array<chunksize=(1032, 2), meta=np.ndarray>
* z_t (z_t) float32 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
Dimensions without coordinates: nlat, nlon, d2
Data variables:
SALT (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
TEMP (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
Attributes: (12/19)
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/ea...
calendar: All years have exactly 365 days.
cell_methods: cell_methods = time: mean ==> the vari...
contents: Diagnostic and Prognostic Variables
nco_openmp_thread_number: 1
nsteps_total: 750
... ...
intake_esm_attrs:vertical_levels: 60.0
intake_esm_attrs:spatial_domain: global_ocean
intake_esm_attrs:start_time: 1920-01-16 12:00:00
intake_esm_attrs:end_time: 2005-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: ocn.20C.monthly,
'ocn.RCP85.monthly': <xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, z_t: 60, nlat: 384, nlon: 320, d2: 2)
Coordinates:
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bound (time, d2) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
* z_t (z_t) float32 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
Dimensions without coordinates: nlat, nlon, d2
Data variables:
SALT (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
TEMP (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
Attributes: (12/21)
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/ea...
NCO: 4.3.4
calendar: All years have exactly 365 days.
cell_methods: cell_methods = time: mean ==> the vari...
contents: Diagnostic and Prognostic Variables
nco_openmp_thread_number: 1
... ...
intake_esm_attrs:vertical_levels: 60.0
intake_esm_attrs:spatial_domain: global_ocean
intake_esm_attrs:start_time: 2006-01-16 12:00:00
intake_esm_attrs:end_time: 2100-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: ocn.RCP85.monthly,
'atm.RCP85.monthly': <xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/21)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
host: tcs-f02n07
important_note: This data is part of the project 'Blin...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.105.cam.i....
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 2006-01-16 12:00:00
intake_esm_attrs:end_time: 2100-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.RCP85.monthly,
'ocn.HIST.monthly': <xarray.Dataset>
Dimensions: (time: 840, z_t: 60, nlat: 384, nlon: 320, d2: 2)
Coordinates:
member_id int64 1
* time (time) object 1850-01-16 12:00:00 ... 1919-12-16 12:00:00
time_bound (time, d2) object dask.array<chunksize=(840, 2), meta=np.ndarray>
* z_t (z_t) float32 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
Dimensions without coordinates: nlat, nlon, d2
Data variables:
SALT (time, z_t, nlat, nlon) float32 dask.array<chunksize=(6, 60, 384, 320), meta=np.ndarray>
TEMP (time, z_t, nlat, nlon) float32 dask.array<chunksize=(6, 60, 384, 320), meta=np.ndarray>
Attributes: (12/19)
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/ea...
calendar: All years have exactly 365 days.
cell_methods: cell_methods = time: mean ==> the vari...
contents: Diagnostic and Prognostic Variables
nco_openmp_thread_number: 1
nsteps_total: 750
... ...
intake_esm_attrs:vertical_levels: 60.0
intake_esm_attrs:spatial_domain: global_ocean
intake_esm_attrs:start_time: 1850-01-16 12:00:00
intake_esm_attrs:end_time: 1919-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: ocn.HIST.monthly,
'atm.HIST.monthly': <xarray.Dataset>
Dimensions: (time: 840, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
member_id int64 1
* time (time) object 1850-01-16 12:00:00 ... 1919-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(840, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (time, lev, lat, lon) float32 dask.array<chunksize=(18, 30, 192, 288), meta=np.ndarray>
T (time, lev, lat, lon) float32 dask.array<chunksize=(18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/20)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
important_note: This data is part of the project 'Blin...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.001.cam.i....
logname: mudryk
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 1850-01-16 12:00:00
intake_esm_attrs:end_time: 1919-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.HIST.monthly,
'atm.20C.monthly': <xarray.Dataset>
Dimensions: (member_id: 40, time: 1032, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 1920-01-16 12:00:00 ... 2005-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1032, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/20)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
important_note: This data is part of the project 'Blin...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.001.cam.i....
logname: mudryk
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 1920-01-16 12:00:00
intake_esm_attrs:end_time: 2005-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.20C.monthly,
'ocn.CTRL.monthly': <xarray.Dataset>
Dimensions: (member_id: 1, time: 21612, z_t: 60, nlat: 384, nlon: 320, d2: 2)
Coordinates:
* member_id (member_id) int64 1
* time (time) object 0400-01-16 12:00:00 ... 2200-12-16 12:00:00
time_bound (time, d2) object dask.array<chunksize=(10806, 2), meta=np.ndarray>
* z_t (z_t) float32 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
Dimensions without coordinates: nlat, nlon, d2
Data variables:
SALT (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
TEMP (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
Attributes: (12/20)
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/ea...
NCO: 4.3.4
calendar: All years have exactly 365 days.
cell_methods: cell_methods = time: mean ==> the vari...
contents: Diagnostic and Prognostic Variables
nco_openmp_thread_number: 1
... ...
intake_esm_attrs:vertical_levels: 60.0
intake_esm_attrs:spatial_domain: global_ocean
intake_esm_attrs:start_time: 0400-01-16 12:00:00
intake_esm_attrs:end_time: 2200-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: ocn.CTRL.monthly,
'atm.CTRL.monthly': <xarray.Dataset>
Dimensions: (member_id: 1, time: 21612, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1
* time (time) object 0400-01-16 12:00:00 ... 2200-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(10806, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/20)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
case: b.e11.B1850C5CN.f09_g16.005
initial_file: /glade/p/cesm/cseg//inputdata/atm/cam/...
logname: mai
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 0400-01-16 12:00:00
intake_esm_attrs:end_time: 2200-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.CTRL.monthly}
How do we work with dictionaries of datasets?
The result of .to_dataset_dict() is a dictionary of datasets, organized via key:xarray.Dataset
For example, we see that our dictionary of datasets has two keys, corresponding to atmospheric, and oceanic components respectively
dsets.keys()
dict_keys(['ocn.20C.monthly', 'ocn.RCP85.monthly', 'atm.RCP85.monthly', 'ocn.HIST.monthly', 'atm.HIST.monthly', 'atm.20C.monthly', 'ocn.CTRL.monthly', 'atm.CTRL.monthly'])
We can access the xarray.Datasets within our dictionary by using these keys! For example, let’s take a look at the atmospheric data.
atmosphere_dset = dsets['atm.RCP85.monthly']
atmosphere_dset
<xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/21)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
host: tcs-f02n07
important_note: This data is part of the project 'Blin...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.105.cam.i....
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 2006-01-16 12:00:00
intake_esm_attrs:end_time: 2100-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.RCP85.monthlyOur dataset has both temperature (T), and specific humidity (Q) within it! We can follow the same process for the ocean data…
ocean_dset = dsets['ocn.RCP85.monthly']
ocean_dset
<xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, z_t: 60, nlat: 384, nlon: 320, d2: 2)
Coordinates:
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bound (time, d2) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
* z_t (z_t) float32 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
Dimensions without coordinates: nlat, nlon, d2
Data variables:
SALT (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
TEMP (member_id, time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 6, 60, 384, 320), meta=np.ndarray>
Attributes: (12/21)
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/ea...
NCO: 4.3.4
calendar: All years have exactly 365 days.
cell_methods: cell_methods = time: mean ==> the vari...
contents: Diagnostic and Prognostic Variables
nco_openmp_thread_number: 1
... ...
intake_esm_attrs:vertical_levels: 60.0
intake_esm_attrs:spatial_domain: global_ocean
intake_esm_attrs:start_time: 2006-01-16 12:00:00
intake_esm_attrs:end_time: 2100-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: ocn.RCP85.monthlyHow do I write analysis code to work with Intake-ESM?
What if we wanted to look at a timeseries of atmospheric data, including both the historical and future scenarios… We need to follow the same few steps
Read in the catalog
Search for our data using
.search()Load in the data using
.to_dataset_dict()
Search for Monthly Atmospheric Data
We restrict the search to atmospheric temperature and specific humidity
data_catalog_subset = data_catalog.search(component='atm',
variable=['T', 'Q'],
frequency='monthly',
experiment=['RCP85', '20C'])
data_catalog_subset.df
| variable | long_name | component | experiment | frequency | vertical_levels | spatial_domain | units | start_time | end_time | path | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Q | specific humidity | atm | 20C | monthly | 30.0 | global | kg/kg | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-20C-Q.zarr |
| 1 | T | temperature | atm | 20C | monthly | 30.0 | global | K | 1920-01-16 12:00:00 | 2005-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-20C-T.zarr |
| 2 | Q | specific humidity | atm | RCP85 | monthly | 30.0 | global | kg/kg | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-Q... |
| 3 | T | temperature | atm | RCP85 | monthly | 30.0 | global | K | 2006-01-16 12:00:00 | 2100-12-16 12:00:00 | s3://ncar-cesm-lens/atm/monthly/cesmLE-RCP85-T... |
Load the Data using .to_dataset_dict()
Similar to before, we load in our dictionary of datasets!
dsets = data_catalog_subset.to_dataset_dict(storage_options={'anon':True})
dsets
--> The keys in the returned dictionary of datasets are constructed as follows:
'component.experiment.frequency'
{'atm.RCP85.monthly': <xarray.Dataset>
Dimensions: (member_id: 40, time: 1140, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 2006-01-16 12:00:00 ... 2100-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1140, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/21)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
host: tcs-f02n07
important_note: This data is part of the project 'Blin...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.105.cam.i....
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 2006-01-16 12:00:00
intake_esm_attrs:end_time: 2100-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.RCP85.monthly,
'atm.20C.monthly': <xarray.Dataset>
Dimensions: (member_id: 40, time: 1032, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 1920-01-16 12:00:00 ... 2005-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1032, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/20)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
important_note: This data is part of the project 'Blin...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.001.cam.i....
logname: mudryk
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 1920-01-16 12:00:00
intake_esm_attrs:end_time: 2005-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.20C.monthly}
Build a Function to Operator on a Single Dataset
A good practice is to write a function which works with a single dataset. Let’s work on an example.
historical_ds = dsets['atm.20C.monthly']
historical_ds
<xarray.Dataset>
Dimensions: (member_id: 40, time: 1032, lev: 30, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* member_id (member_id) int64 1 2 3 4 5 6 7 8 ... 34 35 101 102 103 104 105
* time (time) object 1920-01-16 12:00:00 ... 2005-12-16 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1032, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
Q (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
T (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 18, 30, 192, 288), meta=np.ndarray>
Attributes: (12/20)
Conventions: CF-1.0
NCO: 4.3.4
Version: $Name$
important_note: This data is part of the project 'Blin...
initial_file: b.e11.B20TRC5CNBDRD.f09_g16.001.cam.i....
logname: mudryk
... ...
intake_esm_attrs:vertical_levels: 30.0
intake_esm_attrs:spatial_domain: global
intake_esm_attrs:start_time: 1920-01-16 12:00:00
intake_esm_attrs:end_time: 2005-12-16 12:00:00
intake_esm_attrs:_data_format_: zarr
intake_esm_dataset_key: atm.20C.monthlyLet’s write a simple function to subset for the lowest level (.isel(lev=-1)), and select a lat/lon point of our choosing.
We can do this with only this with the following syntax:
lat = 40.0150
lon = 105.2705
variable = 'T'
ds = historical_ds.isel(member_id=0, lev=-1).sel(lat=lat, lon=lon, method='nearest')[variable]
ds.isel(time=range(10)).plot()
[<matplotlib.lines.Line2D at 0x164f78610>]
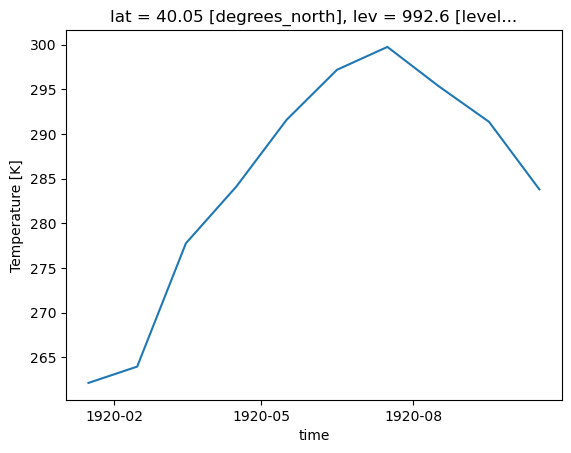
As a function, this would accept a dataset, with a few parameters. We will also subset for the first 5 years (range(60)) for the sake of time.
def plot_point_timeseries(ds, variable, lat=40.015, lon = 105.2705):
ds_subset = ds.isel(member_id=0, lev=-1, time=range(60)).sel(lat=lat, lon=lon, method='nearest')[variable]
ds_subset['time'] = ds_subset.indexes['time'].to_datetimeindex()
return ds_subset.plot(figsize=(10,8))
plot_point_timeseries(historical_ds, variable='Q');
/Users/mgrover/miniforge3/envs/mscar-python-tutorial-dev/lib/python3.10/site-packages/xarray/coding/times.py:360: FutureWarning: Index.ravel returning ndarray is deprecated; in a future version this will return a view on self.
sample = dates.ravel()[0]
/var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/ipykernel_84367/3553229457.py:3: RuntimeWarning: Converting a CFTimeIndex with dates from a non-standard calendar, 'noleap', to a pandas.DatetimeIndex, which uses dates from the standard calendar. This may lead to subtle errors in operations that depend on the length of time between dates.
ds_subset['time'] = ds_subset.indexes['time'].to_datetimeindex()
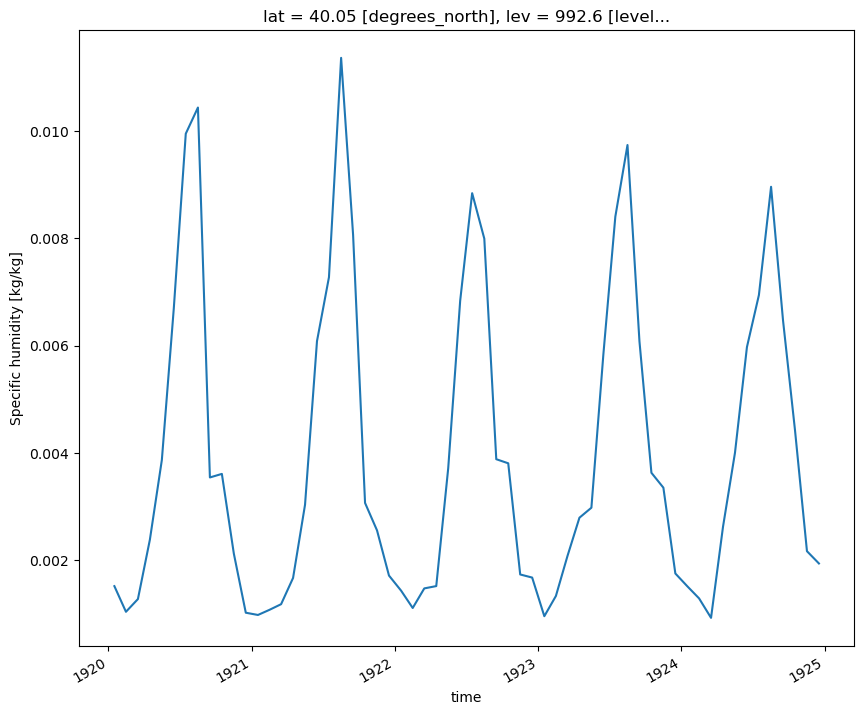
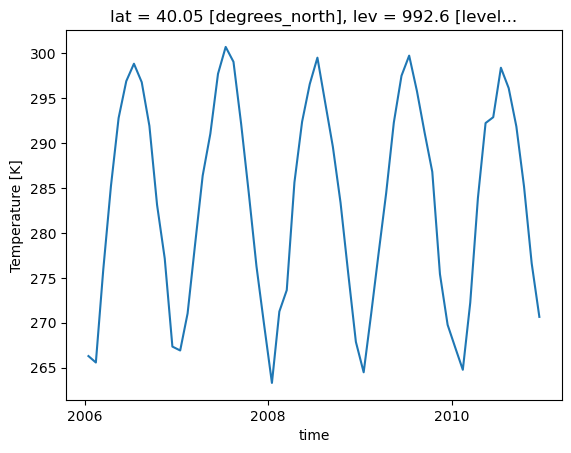
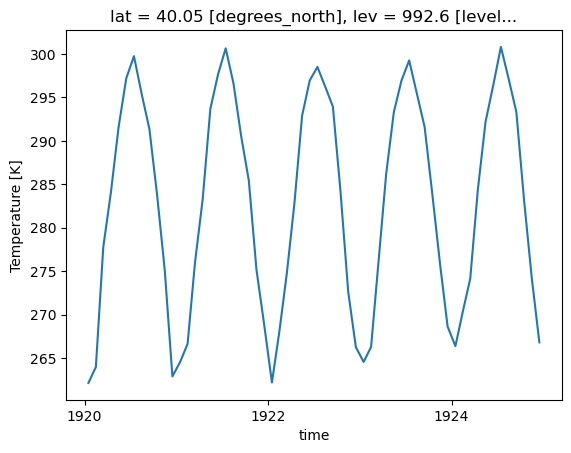
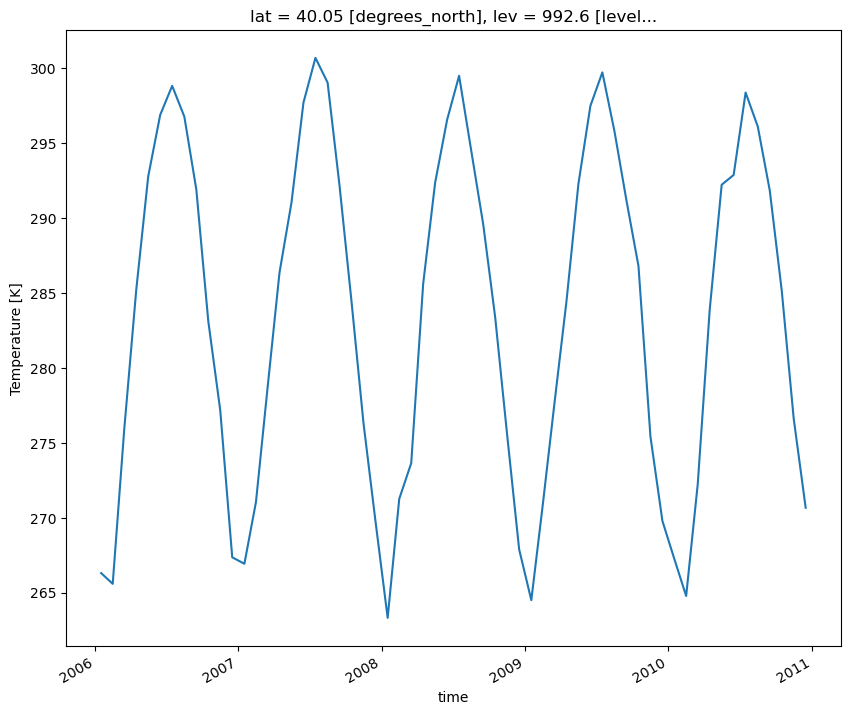
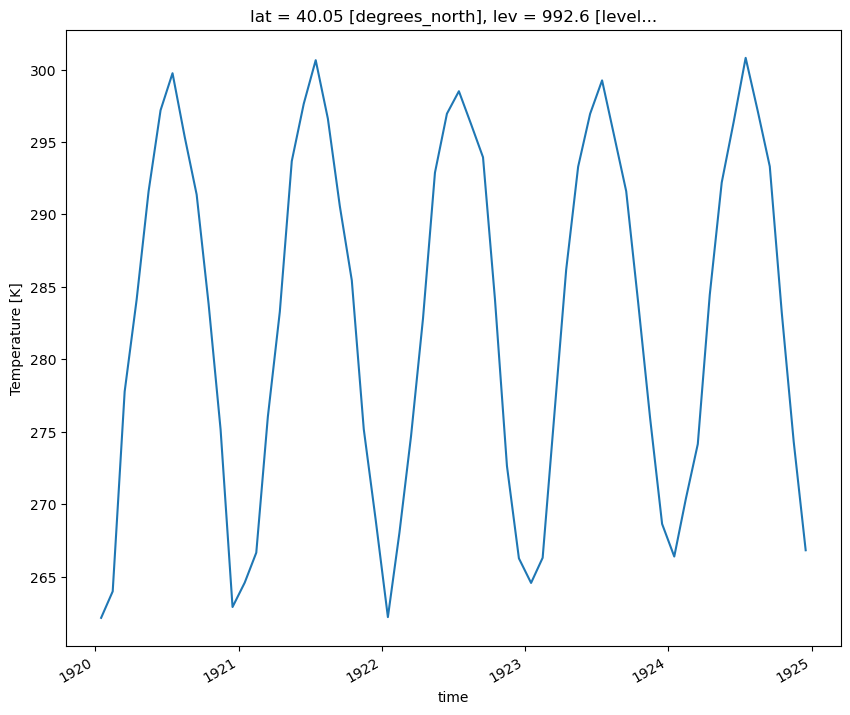
Loop through the Dictionary of Datasets to Apply this Function
# Loop through the different keys in the dictionary of datasets
for key in dsets.keys():
plot = plot_point_timeseries(dsets[key], variable='T')
plt.show()
plt.close()
/Users/mgrover/miniforge3/envs/mscar-python-tutorial-dev/lib/python3.10/site-packages/xarray/coding/times.py:360: FutureWarning: Index.ravel returning ndarray is deprecated; in a future version this will return a view on self.
sample = dates.ravel()[0]
/var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/ipykernel_84367/3553229457.py:3: RuntimeWarning: Converting a CFTimeIndex with dates from a non-standard calendar, 'noleap', to a pandas.DatetimeIndex, which uses dates from the standard calendar. This may lead to subtle errors in operations that depend on the length of time between dates.
ds_subset['time'] = ds_subset.indexes['time'].to_datetimeindex()
/Users/mgrover/miniforge3/envs/mscar-python-tutorial-dev/lib/python3.10/site-packages/xarray/coding/times.py:360: FutureWarning: Index.ravel returning ndarray is deprecated; in a future version this will return a view on self.
sample = dates.ravel()[0]
/var/folders/bw/c9j8z20x45s2y20vv6528qjc0000gq/T/ipykernel_84367/3553229457.py:3: RuntimeWarning: Converting a CFTimeIndex with dates from a non-standard calendar, 'noleap', to a pandas.DatetimeIndex, which uses dates from the standard calendar. This may lead to subtle errors in operations that depend on the length of time between dates.
ds_subset['time'] = ds_subset.indexes['time'].to_datetimeindex()
Use Datatree instead
dt = data_catalog_subset.to_datatree(storage_options={"anon":True})
dt
--> The keys in the returned dictionary of datasets are constructed as follows:
'component/experiment/frequency'
<xarray.DatasetView>
Dimensions: ()
Data variables:
*empty*In a similar fashion to what we used before, we can apply functions that work at the dataset level across our tree of datasets!
timeseries = dt.sel(lat = 40.0150, lon = 105.2705, method='nearest').isel(member_id=0, lev=-1, time=range(60),)
for model_name, model in timeseries.children.items():
try:
timeseries[model_name].ds.T.plot()
plt.show()
plt.close()
except AttributeError:
pass